2017-08-17 14:09:02
Themes
- Is there a reproducibility crisis?
- What is reproducible psychological science?
- How can R make my science more transparent, open, and reproducible?
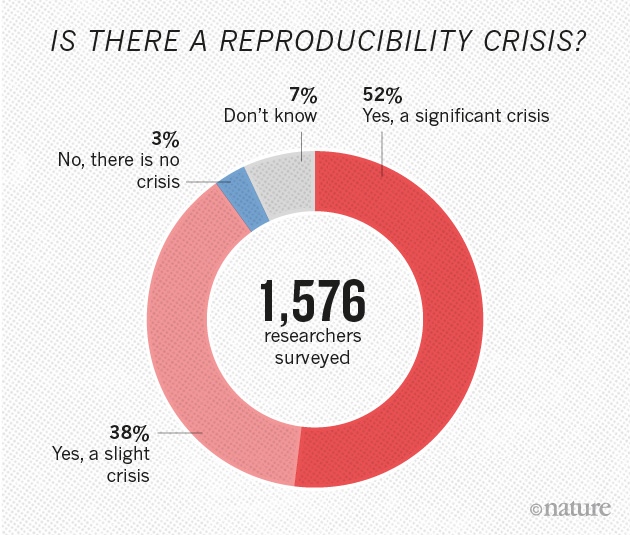
Is there a reproducibility crisis?
- Yes, a significant crisis
- Yes, a slight crisis
- No crisis
- Don't know
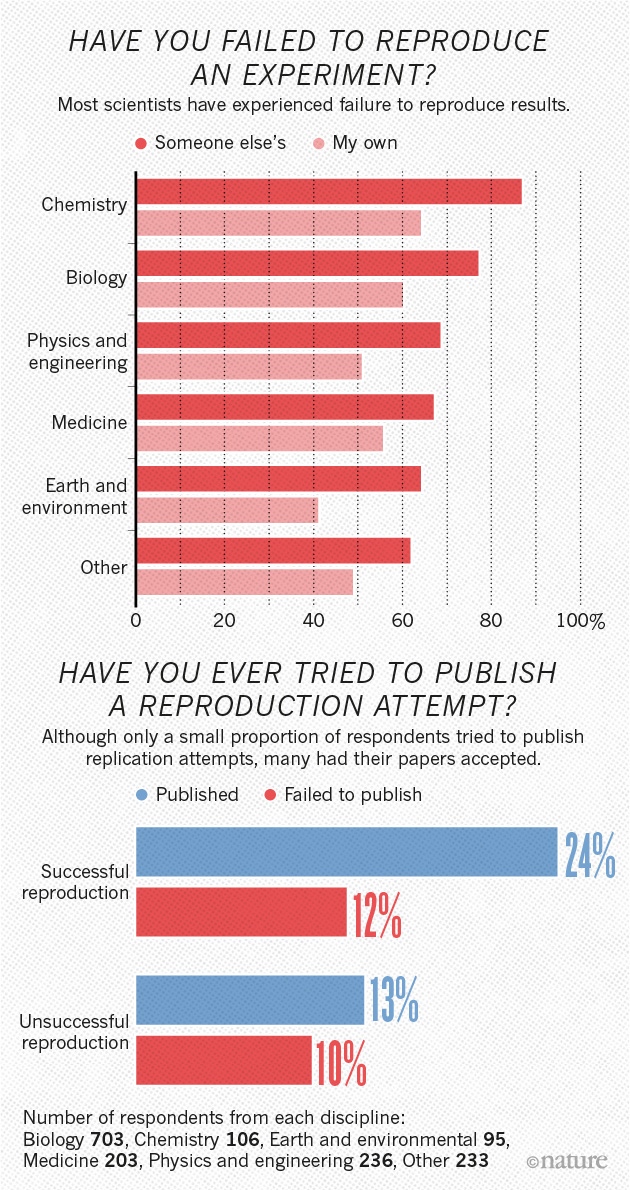
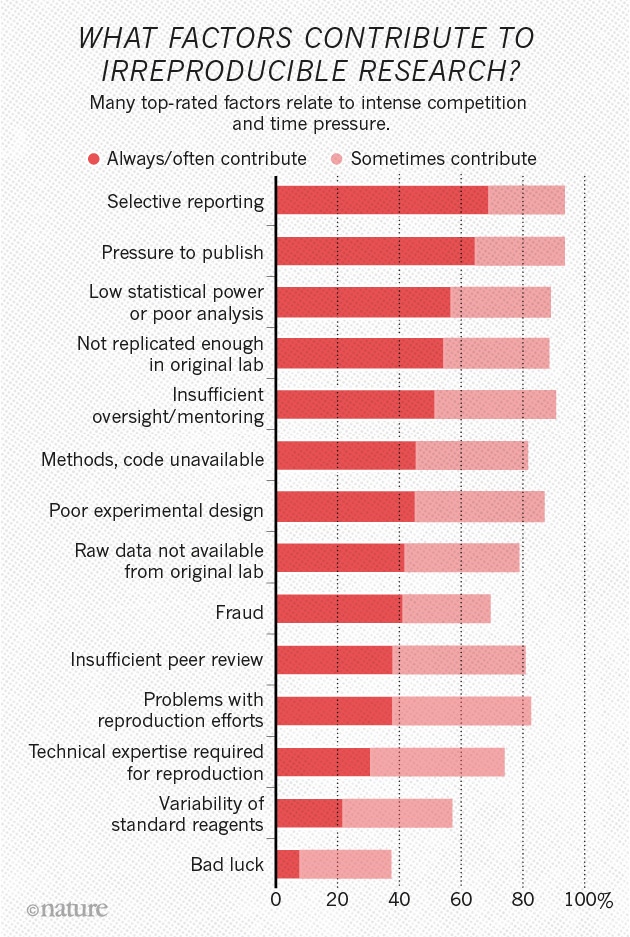
Not just in psychology

(Munafò et al. 2017) manifesto
What am I trying to reproduce?
- My own workflow
- Data collection
- Cleaning
- Visualization
- Analysis
- Reporting
- Manuscript generation?
- "Hit by a truck" scenario
Reproducible workflows
- Scripted, automated = minimize human-dependent steps.
- Well-documented
- Be kind to your future (forgetful) self
- Transparent to me & colleagues == transparent to others
Using R for reproducible workflows
- Option 1: All commands in an R script: e.g.,
project_analysis.R - Option 2a: Mix R code, output, comments in an R Markdown document
- Option 2b: Use R scripts with some special formatting, (more info).
Example 1
# Import data # Clean data # Visualize data # Analyze data # Report findings
# Import data
my_data <- read.csv("path/2/data_file.csv")
# Clean data
my_data$gender <- tolower(my_data$gender) # make lower case
...
Make script that calls sequence of R commands or functions
# Import data
source("R/Import_data.R") # source() runs scripts, loads functions
# Clean data
source("R/Clean_data.R")
# Visualize data
source("R/Visualize_data.R")
...
Strengths & Weaknesses
- R commands in files that can be re-run
- Separate pieces of workflow kept separate
- "Master.R" script that can be run to regenerate full sequence of results
- Error in raw data file?
- No problem; fix and re-run "Master.R"
- How to save results or share with collaborators?
Example 2 - R Markdown
- James' R commands from Day 1: Raw R script (.R)
- Converted to R Markdown
- Output as | HTML notebook | HTML Slides | PDF | docx |
Structure of an R Markdown .Rmd file
- header info in YAML Ain't Markup Language (YAML) format
- Markdown for formating text (headers, boldface/italics,
code, bulleted or numbered lists, web links, etc. - R code "chunks"
One R to rule them all and in the console bind them…
- One file, many possible outputs
- pdf_document, word_document, or github_document
- ioslides_presentation for HTML slide show
- Cool interactive web-app like Dan's tutorial
- Web sites like the one for this bootcamp, blogs, even books
Your turn
- Open "File/New File/R Notebook"
- Change
title: "R Notebook"to something else, liketitle: "Rick's R Notebook" - Save the file (default name is
Untitled) with an.Rmdextension. - Look at the
*.Rmdcode. - Look at the
*.nb.htmlfile in a browser.
Things to try if you like
# Big idea ## Smaller idea in service of bigger - Supporting point - Another suppporting point 1. an enumerated **bold** point 1. an enumerated *italicized* point - a [link](http://psu-psychology.github.io/r-bootcamp) to this bootcamp - an image:  - an equation: $e = mc^2$
Big idea
Smaller idea in service of bigger
- Supporting point
- Another suppporting point
- a bold point
- an italicized point
- a link to this bootcamp
- an image:
- an equation: \(e = mc^2\)
Let's try it with some data
One file, many output options
- 'Default' for the file:
rmarkdown::render("talks/bootcamp-survey.Rmd") - PDF document:
rmarkdown::render('talks/bootcamp-survey.Rmd', output_format = "pdf_document") - Word document:
rmarkdown::render('talks/bootcamp-survey.Rmd', output_format = "word_document")
- HTML slides:
rmarkdown::render('talks/bootcamp-survey.Rmd', output_format = "ioslides_presentation") - Multiple outputs:
rmarkdown::render('talks/bootcamp-survey.Rmd', output_format = c("pdf_document", "word_document", "github_document", "ioslides_presentation")
Key points
- Use R scripts to capture & reproduce workflows and/or
- Use R Markdown files for documents, reports, presentations.
- One or more output formats from the same file.
- Analysis/lab notebook.
- Use R scripts or functions to automate different pieces of the pipeline.
- Make README files to explain how to put pieces together.

Toward a reproducible psychological science…
- Transparent, reproducible, open workflows pre-publication
- Openly shared materials + data + code
- (Munafò et al. 2017): reproducible practices across the workflow
- Where to share and when? Lots of options. Let's talk.
- (Gilmore and Adolph 2017): video and reproducibility
Advanced topics
- Write papers in R Markdown using
papaja - Use R Studio projects
- Version control with git and GitHub
- Scriptable analysis workflows
- Reports for each participant, e.g. PEEP-II project
- This bootcamp's
Make_site.R
- Web sites, blogs, (even books) with R Markdown
R Studio Projects
- Keep files, settings, organized
- Easy to switch between projects
- Reduces mental effort (what directory am I in?)
- Integrates with version control (e.g., GitHub)
Version control
- Keep track of your past
- Back to the Future
- git: a system for software version control
- GitHub: a website for managing projects that use git
My GitHub workflow
- Create a repo on GitHub
- Copy repo URL
File/New Project.../- Version Control, Git
- Paste repo URL
- Select local name for repo and directory where it lives.
- Open project within R Studio
File/Open Project... - Commit early & often
Scripting the pipeline
# Get_bootcamp_googlesheet.R # # Script to authenticate to Google, extract R bootcamp survey data library(googlesheets) library(tidyverse) survey_url <- "https://docs.google.com/spreadsheets/d/1Ay56u6g4jyEEdlmV2NHxTLBlcjI2gHavta-Ik0kGrpg/edit?usp=sharing" bootcamp_by_url <- survey_url %>% extract_key_from_url() %>% gs_key() bootcamp_sheets <- gs_ws_ls(bootcamp_by_url)
boot_data <- bootcamp_by_url %>%
gs_read(bootcamp_sheets[1])
names(boot_data) <- c("Timestamp",
"R_exp",
"GoT",
"Age_yrs",
"Sleep_hrs",
"Fav_date",
"Tidy_data")
write_csv(boot_data, path = "data/survey.csv")
# Update_survey.R
#
# Updates Googlesheet survey data and generates new R Markdown report
#
source("R/Get_bootcamp_googlesheet.R")
rmarkdown::render("talks/bootcamp-survey.Rmd",
output_format = c("github_document",
"pdf_document",
"word_document",
"ioslides_presentation"))
Web sites
_site.yml: site configuration parametersindex.Rmd: home page for site- other
*.Rmdfiles: other pages - other directories for files
rmarkdown::render_site()- GitHub pages or other web site hosting service
Learn from my mistakes
- Script everything you possibly can
- If you have to repeat something, make a function or write a parameterized script
- Document all the time
- Comments in code
- Update README files
- Don't be afraid to ask
- Don't be afraid to work in the open
- Learn from others
- Just do it!
References
Gilmore, Rick O, and Karen E Adolph. 2017. “Video Can Make Behavioural Science More Reproducible.” Nature Human Behavior 1 (12~jun). doi:10.1038/s41562-017-0128.
Munafò, Marcus R, Brian A Nosek, Dorothy V M Bishop, Katherine S Button, Christopher D Chambers, Nathalie Percie du Sert, Uri Simonsohn, Eric-Jan Wagenmakers, Jennifer J Ware, and John P A Ioannidis. 2017. “A Manifesto for Reproducible Science.” Nature Human Behaviour 1 (10~jan): 0021. doi:10.1038/s41562-016-0021.